Word repetition memory¶
This builds on the P50 and P300 example.
Procedure: A category prompt is presented, e.g. “large river” or “part of a foot”. After a 3 s pause the probe word is presented, e.g., “toe” followed by a verbal Yes or No indicating whether the word fit the category.
Design
half the probe words are congruous with the prompt, half are incongruous
the prompt-probe word pairs are presented one, two, or three times
repeated prompt-probe word pairs are presented at short and long lags
Event code tagging
each probe word has a unique integer event code
the codemap patterns tag individual words for item analysis
event tags include the prompt and probe word stimulus
[1]:
import os
import sys
from pathlib import Path
import re
import numpy as np
import pandas as pd
import mkpy
import spudtr
from matplotlib import pyplot as plt
from mkpy import mkh5
from spudtr import epf
# path wrangling for nbsphinx
if "MDE_HOME" in os.environ.keys():
MDE_HOME = Path(os.environ["MDE_HOME"])
else:
from conf import MDE_HOME
DOCS_DATA = MDE_HOME / "docs/_data"
print(os.environ["CONDA_DEFAULT_ENV"])
print(sys.version)
for pkg in [np, pd, mkpy, spudtr]:
print(pkg.__name__, pkg.__version__, pkg.__file__)
mkconda_dev_py39_053022
3.9.13 | packaged by conda-forge | (main, May 27 2022, 16:56:21)
[GCC 10.3.0]
numpy 1.21.6 /home/turbach/miniconda39/envs/mkconda_dev_py39_053022/lib/python3.9/site-packages/numpy/__init__.py
pandas 1.1.5 /home/turbach/miniconda39/envs/mkconda_dev_py39_053022/lib/python3.9/site-packages/pandas/__init__.py
mkpy 0.2.7 /mnt/cube/home/turbach/TPU_Projects/mkpy/mkpy/__init__.py
spudtr 0.1.0 /home/turbach/miniconda39/envs/mkconda_dev_py39_053022/lib/python3.9/site-packages/spudtr/__init__.py
[2]:
# set filenames
crw = MDE_HOME / "mkdig/sub000wr.crw" # EEG recording
log = MDE_HOME / "mkdig/sub000wr.x.log" # events
yhdr = MDE_HOME / "mkpy/sub000wr.yhdr" # extra header info
# set calibration data filenames
cals_crw = MDE_HOME / "mkdig/sub000c.crw"
cals_log = MDE_HOME / "mkdig/sub000c.x.log"
cals_yhdr = MDE_HOME / "mkpy/sub000c.yhdr"
# HDF5 file with EEG recording, events, and header
wr_h5_f = DOCS_DATA / "sub000wr.h5"
mkh5 EEG data, event code log, header information
[3]:
# convert to HDF5
wr_h5 = mkh5.mkh5(wr_h5_f)
wr_h5.reset_all()
wr_h5.create_mkdata("sub000", crw, log, yhdr)
# add calibration data
wr_h5.append_mkdata("sub000", cals_crw, cals_log, cals_yhdr)
# calibrate
pts, pulse, lo, hi, ccode = 5, 10, -40, 40, 0
wr_h5.calibrate_mkdata(
"sub000", # data group to calibrate with these cal pulses
n_points=pts, # pts to average
cal_size=pulse, # uV
lo_cursor=lo, # lo_cursor ms
hi_cursor=hi, # hi_cursor ms
cal_ccode=ccode, # condition code
)
/mnt/cube/home/turbach/TPU_Projects/mkpy/mkpy/mkh5.py:3666: UserWarning: negative event code(s) found for cal condition code 0 -16384
warnings.warn(msg)
Found cals in /sub000/dblock_2
Calibrating block /sub000/dblock_0 of 3: (311040,)
Calibrating block /sub000/dblock_1 of 3: (334592,)
Calibrating block /sub000/dblock_2 of 3: (28416,)
codemap: single trial item analysis
Simple YAML codemaps are fine for simple categorical designs with a few kinds of codes.
To tag single trials with design categories AND individual item attributes for hundreds of stimuli it may be easier to create (and check) the code map in an .xlsx spreadsheet with the help of formulas and macros.
The complexity of the design (8 conditions_ is multiplied by the number of individual items (216)
There are 1738 individual pattern : tags which an be created, stored, managed, and checked in a spreadsheet like so:
1. get_event_table(codemap)
Scan the event codes for pattern matches and collect the HDF5 index, matching code, and tags
Event table modifications are not needed for this simple design.
[4]:
wr_event_table = wr_h5.get_event_table(MDE_HOME / "mkpy/wr_codemap.xlsx")
/mnt/cube/home/turbach/TPU_Projects/mkpy/mkpy/mkh5.py:1059: UserWarning:
As of mkpy 0.2.0 to match events with a codemap regexp pattern, the
ccode column in wr_codemap.xlsx must also match the log_ccode
in the datablock. If this behavior is not desired, delete or rename
the ccode column in the codemap.
warnings.warn(msg)
searching codes in: sub000/dblock_0
searching codes in: sub000/dblock_1
searching codes in: sub000/dblock_2
inspect the event table
[5]:
print(wr_event_table.shape)
print(wr_event_table.columns)
# select some columns to show
example_columns = [
"dblock_path", "dblock_ticks", "log_evcodes", "log_ccodes", "log_flags",
"regexp", "match_code",
"instrument", "item_id", "probe", "congruity", "repetition",
]
# first few stimulus events
display(wr_event_table[example_columns].head())
# last few calibration pulse events
display(wr_event_table[example_columns].tail())
pd.crosstab(
[
wr_event_table.instrument,
wr_event_table.ccode,
wr_event_table.repetition,
wr_event_table.pres_position,
wr_event_table.word_lag,
],
[
wr_event_table.log_flags
],
margins=True
)
(497, 35)
Index(['data_group', 'dblock_path', 'dblock_tick_idx', 'dblock_ticks',
'crw_ticks', 'raw_evcodes', 'log_evcodes', 'log_ccodes', 'log_flags',
'epoch_match_tick_delta', 'epoch_ticks', 'dblock_srate', 'match_group',
'idx', 'dlim', 'anchor_str', 'match_str', 'anchor_code', 'match_code',
'anchor_tick', 'match_tick', 'anchor_tick_delta', 'is_anchor', 'regexp',
'ccode', 'instrument', 'repetition', 'pres_position', 'word_lag',
'congruity', 'event_code', 'condition_id', 'item_id', 'category',
'probe'],
dtype='object')
| dblock_path | dblock_ticks | log_evcodes | log_ccodes | log_flags | regexp | match_code | instrument | item_id | probe | congruity | repetition | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 144 | sub000/dblock_0 | 219511 | 2009 | 2 | 0 | (#2009) | 2009 | eeg | 9 | bitter | congruent | initial |
| 146 | sub000/dblock_0 | 223966 | 3009 | 2 | 0 | (#3009) | 3009 | eeg | 9 | bitter | congruent | repeated |
| 148 | sub000/dblock_0 | 266277 | 8010 | 2 | 0 | (#8010) | 8010 | eeg | 10 | addict | incongruent | initial |
| 150 | sub000/dblock_0 | 276597 | 9010 | 2 | 0 | (#9010) | 9010 | eeg | 10 | addict | incongruent | repeated |
| 152 | sub000/dblock_0 | 108673 | 1011 | 2 | 0 | (#1011) | 1011 | eeg | 11 | blanket | congruent | initial |
| dblock_path | dblock_ticks | log_evcodes | log_ccodes | log_flags | regexp | match_code | instrument | item_id | probe | congruity | repetition | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1068 | sub000/dblock_2 | 27315 | 4 | 0 | 0 | (#\d+) | 4 | cal | -1 | cal | cal | cal |
| 1069 | sub000/dblock_2 | 27444 | 2 | 0 | 0 | (#\d+) | 2 | cal | -1 | cal | cal | cal |
| 1070 | sub000/dblock_2 | 27573 | 3 | 0 | 0 | (#\d+) | 3 | cal | -1 | cal | cal | cal |
| 1071 | sub000/dblock_2 | 27703 | 4 | 0 | 0 | (#\d+) | 4 | cal | -1 | cal | cal | cal |
| 1072 | sub000/dblock_2 | 27832 | 2 | 0 | 0 | (#\d+) | 2 | cal | -1 | cal | cal | cal |
[5]:
| log_flags | 0 | 32 | 48 | 64 | All | ||||
|---|---|---|---|---|---|---|---|---|---|
| instrument | ccode | repetition | pres_position | word_lag | |||||
| cal | 0 | cal | cal | cal | 208 | 0 | 0 | 1 | 209 |
| eeg | 2 | initial | first | long | 21 | 3 | 0 | 0 | 24 |
| short | 24 | 0 | 0 | 0 | 24 | ||||
| unique | 24 | 0 | 0 | 0 | 24 | ||||
| repeated | second | long | 21 | 3 | 0 | 0 | 24 | ||
| short | 22 | 1 | 1 | 0 | 24 | ||||
| third | long | 21 | 3 | 0 | 0 | 24 | |||
| 3 | initial | first | long | 20 | 4 | 0 | 0 | 24 | |
| short | 22 | 2 | 0 | 0 | 24 | ||||
| unique | 23 | 1 | 0 | 0 | 24 | ||||
| repeated | second | long | 22 | 2 | 0 | 0 | 24 | ||
| short | 21 | 3 | 0 | 0 | 24 | ||||
| third | long | 19 | 5 | 0 | 0 | 24 | |||
| All | 468 | 27 | 1 | 1 | 497 |
2. set_epochs(name, pre, post)
Stash the the event table events and tags with a name and epoch interval boundaries.
[6]:
wr_h5.set_epochs("ms1500", wr_event_table, -750, 750)
Sanitizing event table data types for mkh5 epochs table ...
/mnt/cube/home/turbach/TPU_Projects/mkpy/mkpy/mkh5.py:1515: UserWarning: data error: pre-stimulus interval is out of bounds left ... skipping epoch (288, b'sub000', b'sub000/dblock_2', 0, 26, 26, 2, 2, 0, 64, -187, 375, 250., 1, 0, 1, b'2', b'2', 2, 2, 26, 26, 0, True, b'(#\\d+)', 0, b'cal', b'cal', b'cal', b'cal', b'cal', -1, 0, -1, b'cal', b'cal', 0, 0, 0, 26, -187, 375)
warnings.warn(
3. export_epochs(name, file_format=…)
Export the time-stamped, tagged, fixed-length segments of EEG data for analysis as HDF5, pandas HDF5 or feather
[7]:
wr_epochs_f = DOCS_DATA / "sub000wr.ms1500.epochs.feather"
wr_h5.export_epochs("ms1500", wr_epochs_f, file_format="feather")
analyze the epochs
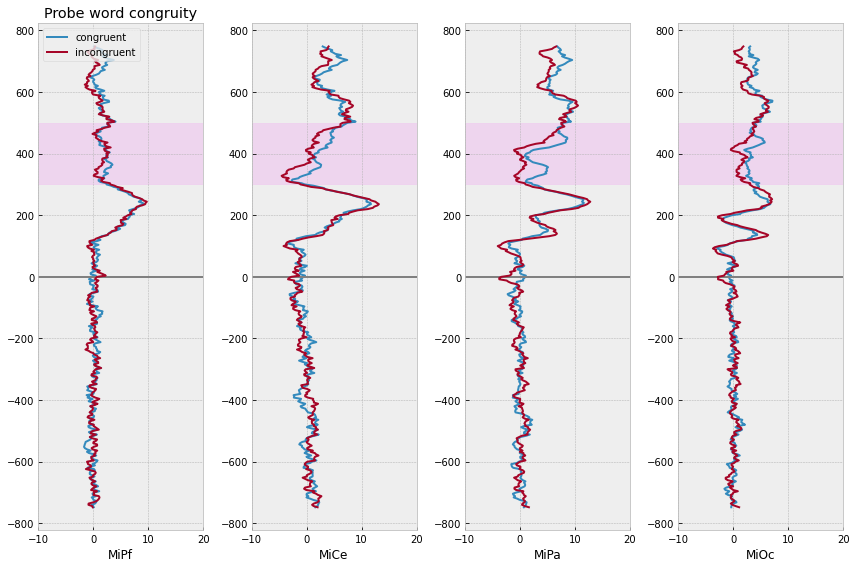
Is there a difference between congruous and incongrous probes?
If so, is the effect the same for first and repeated presentations?
[8]:
from matplotlib import pyplot as plt
from spudtr import epf
plt.style.use('bmh')
wr_epochs = pd.read_feather(wr_epochs_f)
# sanitize Epoch_idx for pandas index
wr_epochs['epoch_id'] = wr_epochs['epoch_id'].astype('int')
# 26 cap + EOG and A2 channel labels
wr_chans = [col for col in wr_epochs.columns if re.match(r"^[MLRAHlr]\w{1,3}$", col)]
print(f"n channels: {len(wr_chans)}\nchannels:{wr_chans}")
# lookup the epochs with stim events at time == 0) and clean artifact flags (= 0)
good_epoch_ids = wr_epochs.query("match_time == 0 and log_flags == 0").epoch_id
# select just the good epochs
wr_epochs = wr_epochs.query("epoch_id in @good_epoch_ids and ccode > 0").copy()
# check the good event counts after dropping artifacts
wr_good_events = wr_epochs.query("match_time == 0")
print("After excluding EEG artifacts")
pd.crosstab(
[
wr_good_events.instrument,
wr_good_events.ccode,
wr_good_events.pres_position,
wr_good_events.word_lag,
],
[
wr_good_events.log_flags
],
margins=True
)
n channels: 32
channels:['lle', 'lhz', 'MiPf', 'LLPf', 'RLPf', 'LMPf', 'RMPf', 'LDFr', 'RDFr', 'LLFr', 'RLFr', 'LMFr', 'RMFr', 'LMCe', 'RMCe', 'MiCe', 'MiPa', 'LDCe', 'RDCe', 'LDPa', 'RDPa', 'LMOc', 'RMOc', 'LLTe', 'RLTe', 'LLOc', 'RLOc', 'MiOc', 'A2', 'HEOG', 'rle', 'rhz']
After excluding EEG artifacts
[8]:
| log_flags | 0 | All | |||
|---|---|---|---|---|---|
| instrument | ccode | pres_position | word_lag | ||
| eeg | 2 | first | long | 21 | 21 |
| short | 24 | 24 | |||
| unique | 24 | 24 | |||
| second | long | 21 | 21 | ||
| short | 22 | 22 | |||
| third | long | 21 | 21 | ||
| 3 | first | long | 20 | 20 | |
| short | 22 | 22 | |||
| unique | 23 | 23 | |||
| second | long | 22 | 22 | ||
| short | 21 | 21 | |||
| third | long | 19 | 19 | ||
| All | 260 | 260 |
[9]:
midline = ["MiPf", "MiCe", "MiPa", "MiOc"]
# some factors of interest
midline_epochs = wr_epochs[
["epoch_id", "match_time", "congruity", "repetition", "pres_position", "word_lag"] + midline
]
# center each channel prestimulus
midline_epochs = epf.center_eeg(
midline_epochs,
midline,
-750, 0,
epoch_id="epoch_id",
time="match_time"
)
[10]:
# compute domain average by stim type
midline_erps = midline_epochs.groupby(
["congruity", "match_time"]
).mean().reset_index()
# plot
f, axs = plt.subplots(1, 4, figsize=(12,8))
for rep, erp in midline_erps.groupby(["congruity"]):
for axi, chan in enumerate(midline):
# mark onset
axs[axi].axhline(0, color='gray')
axs[axi].plot(
erp[chan],
erp["match_time"],
label=f"{rep}",
lw=2,
)
axs[axi].set(xlim=(-10, 20), xlabel=chan)
axs[axi].axhspan(300, 500, color="magenta", alpha=.05)
axs[0].legend(loc="upper left")
axs[0].set_title("Probe word congruity")
f.tight_layout()
[11]:
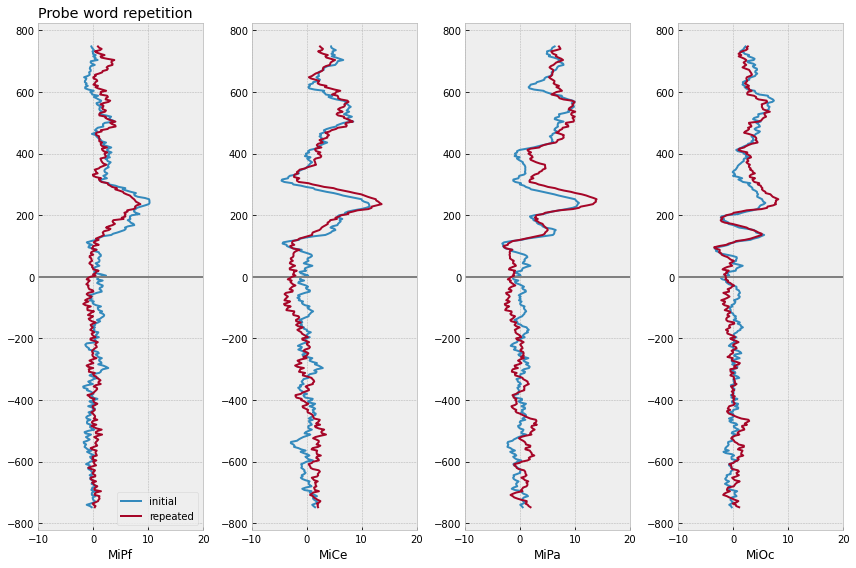
# compute domain average by stim type
midline_erps = midline_epochs.groupby(
["repetition", "match_time"]
).mean().reset_index()
# plot
f, axs = plt.subplots(1, 4, figsize=(12,8))
for rep, erp in midline_erps.groupby(["repetition"]):
for axi, chan in enumerate(midline):
# mark onset
axs[axi].axhline(0, color='gray')
axs[axi].plot(
erp[chan],
erp["match_time"],
label=f"{rep}",
lw=2,
)
axs[axi].set(xlim=(-10, 20), xlabel=chan)
axs[0].legend(loc="lower right")
axs[0].set_title("Probe word repetition", loc='left')
f.tight_layout()
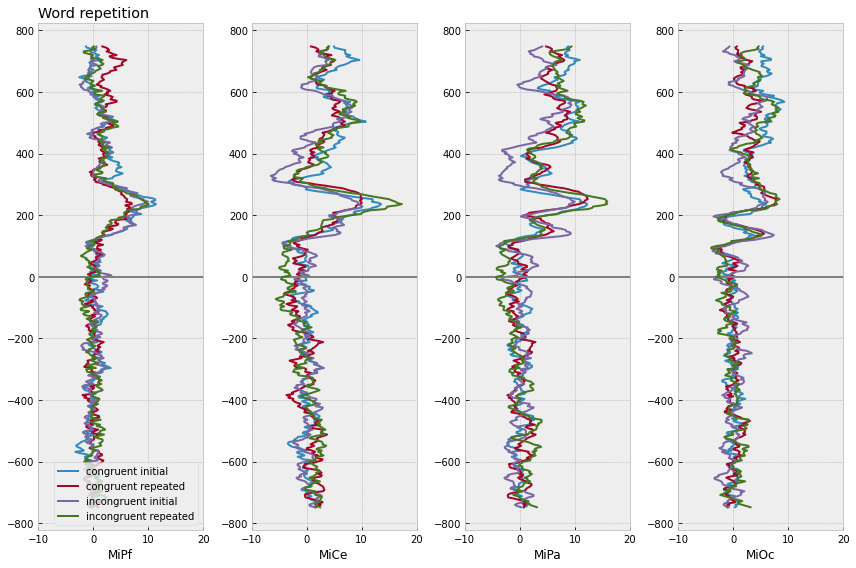
[12]:
midline_erps = midline_epochs.groupby(
["congruity", "repetition", "match_time"]
).mean().reset_index()
# plot
f, axs = plt.subplots(1, 4, figsize=(12,8))
for (congruity, repetition), erp in midline_erps.groupby(['congruity', "repetition"]):
for axi, chan in enumerate(midline):
# mark onset
axs[axi].axhline(0, color='gray')
axs[axi].plot(
erp[chan],
erp["match_time"],
label=f"{congruity} {repetition}",
lw=2,
)
axs[axi].set(xlim=(-10, 20), xlabel=chan)
axs[0].legend(loc="lower right")
axs[0].set_title("Word repetition", loc='left')
f.tight_layout()