Picture memory¶
This builds on the P300 and WR example.
Design * study-test recognition memory * Study phase: 50 pictures were presented for 2s and judged “like” or “dislike”, indicated by a button press * Test phase: The same 50 studied items were presented with 50 distractor items in pseudo-random order and judged “old” or “new”. * The 50 studied and 50 distractor items are counterbalanced across subjects so the individual pictures are studied for half the participants and distractors for the other half.
Automated code tagging * tag individual stimulus items are * stimulus categories * response categories * local (within-trial) stimulus-response contigent tags: “old”, “new”, “hit”, “miss”, “correct rejection”, “false alarm”
Event table modification for custom code tagging * non-local (across-phase) response-contingent tags * separate codemaps and event tables are constructed for the study and test phases * DM analysis: align the study phase items with subsequent test phase “hit”, “miss” responses * Memory x preference: align the test phase items with previous study phase “like”, “dislike” responses
See Appendex at the end of the end of the notebook for source code to generate the code maps.
[1]:
import os
import sys
from pathlib import Path
import re
import numpy as np
import pandas as pd
import mkpy
import spudtr
from matplotlib import pyplot as plt
from mkpy import mkh5
from spudtr import epf
# path wrangling for nbsphinx
if "MDE_HOME" in os.environ.keys():
MDE_HOME = Path(os.environ["MDE_HOME"])
else:
from conf import MDE_HOME
DOCS_DATA = MDE_HOME / "docs/_data"
print(os.environ["CONDA_DEFAULT_ENV"])
print(sys.version)
for pkg in [np, pd, mkpy, spudtr]:
print(pkg.__name__, pkg.__version__, pkg.__file__)
mkconda_dev_py39_053022
3.9.13 | packaged by conda-forge | (main, May 27 2022, 16:56:21)
[GCC 10.3.0]
numpy 1.21.6 /home/turbach/miniconda39/envs/mkconda_dev_py39_053022/lib/python3.9/site-packages/numpy/__init__.py
pandas 1.1.5 /home/turbach/miniconda39/envs/mkconda_dev_py39_053022/lib/python3.9/site-packages/pandas/__init__.py
mkpy 0.2.7 /mnt/cube/home/turbach/TPU_Projects/mkpy/mkpy/__init__.py
spudtr 0.1.0 /home/turbach/miniconda39/envs/mkconda_dev_py39_053022/lib/python3.9/site-packages/spudtr/__init__.py
[2]:
# set filenames
crw = MDE_HOME / "mkdig/sub000pm.crw" # EEG recording
log = MDE_HOME / "mkdig/sub000pm.x.log" # events
yhdr = MDE_HOME / "mkpy/sub000pm.yhdr" # extra header info
# set calibration data filenames
cals_crw = MDE_HOME / "mkdig/sub000c.crw"
cals_log = MDE_HOME / "mkdig/sub000c.x.log"
cals_yhdr = MDE_HOME / "mkpy/sub000c.yhdr"
# HDF5 file with EEG recording, events, and header
pm_h5_f = DOCS_DATA / "sub000pm.h5"
mkh5 EEG data, event code log, header information
[3]:
# convert to HDF5
pm_h5 = mkh5.mkh5(pm_h5_f)
pm_h5.reset_all()
pm_h5.create_mkdata("sub000", crw, log, yhdr)
# add calibration data
pm_h5.append_mkdata("sub000", cals_crw, cals_log, cals_yhdr)
# calibrate
pts, pulse, lo, hi, ccode = 5, 10, -40, 40, 0
pm_h5.calibrate_mkdata(
"sub000", # data group to calibrate with these cal pulses
n_points=pts, # pts to average
cal_size=pulse, # uV
lo_cursor=lo, # lo_cursor ms
hi_cursor=hi, # hi_cursor ms
cal_ccode=ccode, # condition code
)
/mnt/cube/home/turbach/TPU_Projects/mkpy/mkpy/mkh5.py:3666: UserWarning: negative event code(s) found for cal condition code 0 -16384
warnings.warn(msg)
Found cals in /sub000/dblock_3
Calibrating block /sub000/dblock_0 of 4: (95232,)
Calibrating block /sub000/dblock_1 of 4: (34048,)
Calibrating block /sub000/dblock_2 of 4: (139008,)
Calibrating block /sub000/dblock_3 of 4: (28416,)
codemaps: study and test phase generated programmatically from a table of item information
item information
Begin with the item-specific information, gathered somehow.
Here awk extracts a flat text file directly from the actual stimulus presentation files.
condition_id tracks animacy and item_id tracks the item.
jpg is the image file prefix for human readability and scn file tracks the version of the stimulus presentation file which counterbalance the 50 study items so of the 100 test items, across subjects, each individual picture appears equally often as previously presented or not.
[4]:
%%bash
echo "study phase head"
head ${MDE_HOME}/mkpy/pm_item_id_by_scn.tsv
echo "test phase tail"
tail ${MDE_HOME}/mkpy/pm_item_id_by_scn.tsv
study phase head
ccode phase condition_id item_id jpg scn
2 study 2 154 necklace studyp1
2 study 2 107 bell studyp1
2 study 1 146 leopard studyp1
2 study 1 157 peas studyp1
2 study 2 151 moon studyp1
2 study 2 143 jacket studyp1
2 study 2 147 lighter studyp1
2 study 2 141 htarblon studyp1
2 study 2 149 lock studyp1
test phase tail
1 test 1 144 kilerwal testp4
1 test 2 187 table testp4
1 test 4 111 briefcas testp4
1 test 4 197 whistle testp4
1 test 4 200 wrench testp4
1 test 4 152 motrcyle testp4
1 test 2 120 chair testp4
1 test 3 186 swan testp4
1 test 4 167 refridge testp4
1 test 2 194 vacuum testp4
[5]:
# read the item information table
pm_items = pd.read_csv(
MDE_HOME / "mkpy/pm_item_id_by_scn.tsv",
delim_whitespace=True
).query("scn in ['studyp1', 'testp1']")
display(pm_items.head())
display(pm_items.tail())
| ccode | phase | condition_id | item_id | jpg | scn | |
|---|---|---|---|---|---|---|
| 0 | 2 | study | 2 | 154 | necklace | studyp1 |
| 1 | 2 | study | 2 | 107 | bell | studyp1 |
| 2 | 2 | study | 1 | 146 | leopard | studyp1 |
| 3 | 2 | study | 1 | 157 | peas | studyp1 |
| 4 | 2 | study | 2 | 151 | moon | studyp1 |
| ccode | phase | condition_id | item_id | jpg | scn | |
|---|---|---|---|---|---|---|
| 295 | 1 | test | 4 | 198 | wineglas | testp1 |
| 296 | 1 | test | 4 | 125 | crayon | testp1 |
| 297 | 1 | test | 3 | 175 | sheep | testp1 |
| 298 | 1 | test | 4 | 184 | stove | testp1 |
| 299 | 1 | test | 2 | 105 | barn | testp1 |
study phase codemap
define a code pattern: tags template
plug in the individual items
save the result
Note that many-to-many mapping. Many codes (1, 2) are mapped to many tags, e.g., “like”, “dislike”
[6]:
# study phase codemape file name
pm_study_codemap_f = MDE_HOME / "mkpy/pm_study_codemap.tsv"
pm_study_codemap_cols = ["regexp", "study_bin_id", "animacy", "study_response", ] + list(pm_items.columns)
pm_study_codemap = pd.read_csv(pm_study_codemap_f, sep="\t")
display(pm_study_codemap.shape)
display(pm_study_codemap)
(702, 10)
| regexp | study_bin_id | animacy | study_response | ccode | phase | condition_id | item_id | jpg | scn | |
|---|---|---|---|---|---|---|---|---|---|---|
| 0 | (#[1234]) | 0 | cal | cal | 0 | study | 0 | -1 | cal | cal |
| 1 | (#[12]) | 200 | _any | _any | 2 | study | 2 | -1 | _any | _any |
| 2 | (#[1]) 8 (154) 1040 | 2100 | animate | like | 2 | study | 2 | 154 | necklace | studyp1 |
| 3 | (#[1]) 8 1040 (154) | 2101 | animate | like | 2 | study | 2 | 154 | necklace | studyp1 |
| 4 | (#[1]) 1040 8 (154) | 2102 | animate | like | 2 | study | 2 | 154 | necklace | studyp1 |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 697 | (#[2]) 1040 8 (159) | 2202 | inanimate | like | 2 | study | 2 | 159 | pen | studyp1 |
| 698 | (#[2]) 8 (159) 2064 | 2210 | inanimate | dislike | 2 | study | 2 | 159 | pen | studyp1 |
| 699 | (#[2]) 8 2064 (159) | 2211 | inanimate | dislike | 2 | study | 2 | 159 | pen | studyp1 |
| 700 | (#[2]) 2064 8 (159) | 2212 | inanimate | dislike | 2 | study | 2 | 159 | pen | studyp1 |
| 701 | (#[2]) 8 (159) (?!(1040|2064)) | 2203 | inanimate | no_response | 2 | study | 2 | 159 | pen | studyp1 |
702 rows × 10 columns
1. study phase get_event_table(codemap)
[7]:
pm_study_event_table = pm_h5.get_event_table(pm_study_codemap_f)
searching codes in: sub000/dblock_0
/mnt/cube/home/turbach/TPU_Projects/mkpy/mkpy/mkh5.py:1059: UserWarning:
As of mkpy 0.2.0 to match events with a codemap regexp pattern, the
ccode column in pm_study_codemap.tsv must also match the log_ccode
in the datablock. If this behavior is not desired, delete or rename
the ccode column in the codemap.
warnings.warn(msg)
searching codes in: sub000/dblock_1
searching codes in: sub000/dblock_2
searching codes in: sub000/dblock_3
inspect the study phase event table
[8]:
print("study phase (shape):", pm_study_event_table.shape)
print("study phase columns:", pm_study_event_table.columns.to_list())
# select some columns to show
example_columns = [
"dblock_path", "dblock_ticks", "log_evcodes", "log_ccodes", "log_flags",
"regexp", "match_code",
"phase", "study_bin_id", "study_response",
]
# first few stimulus events, coded for response
display(pm_study_event_table[example_columns].query("study_bin_id > 2000").head())
for is_anchor in [True, False]:
print("is_anchor: ", is_anchor)
events = pm_study_event_table.query("is_anchor == @is_anchor ")
print(events.shape)
display(
pd.crosstab(
[
events.data_group,
events.ccode,
events.study_bin_id,
events.study_response
],
[
events.log_flags
],
margins=True
)
)
study phase (shape): (359, 33)
study phase columns: ['data_group', 'dblock_path', 'dblock_tick_idx', 'dblock_ticks', 'crw_ticks', 'raw_evcodes', 'log_evcodes', 'log_ccodes', 'log_flags', 'epoch_match_tick_delta', 'epoch_ticks', 'dblock_srate', 'match_group', 'idx', 'dlim', 'anchor_str', 'match_str', 'anchor_code', 'match_code', 'anchor_tick', 'match_tick', 'anchor_tick_delta', 'is_anchor', 'regexp', 'study_bin_id', 'animacy', 'study_response', 'ccode', 'phase', 'condition_id', 'item_id', 'jpg', 'scn']
| dblock_path | dblock_ticks | log_evcodes | log_ccodes | log_flags | regexp | match_code | phase | study_bin_id | study_response | |
|---|---|---|---|---|---|---|---|---|---|---|
| 100 | sub000/dblock_0 | 834 | 2 | 2 | 32 | (#[2]) 8 (154) 1040 | 2 | study | 2200 | like |
| 101 | sub000/dblock_0 | 1410 | 154 | 2 | 0 | (#[2]) 8 (154) 1040 | 154 | study | 2200 | like |
| 102 | sub000/dblock_0 | 3390 | 2 | 2 | 0 | (#[2]) 8 (107) 1040 | 2 | study | 2200 | like |
| 103 | sub000/dblock_0 | 3969 | 107 | 2 | 0 | (#[2]) 8 (107) 1040 | 107 | study | 2200 | like |
| 104 | sub000/dblock_0 | 5532 | 1 | 2 | 0 | (#[1]) 8 (146) 1040 | 1 | study | 2100 | like |
is_anchor: True
(309, 33)
| log_flags | 0 | 32 | 64 | All | |||
|---|---|---|---|---|---|---|---|
| data_group | ccode | study_bin_id | study_response | ||||
| sub000 | 0 | 0 | cal | 208 | 0 | 1 | 209 |
| 2 | 200 | _any | 48 | 2 | 0 | 50 | |
| 2100 | like | 9 | 0 | 0 | 9 | ||
| 2101 | like | 1 | 0 | 0 | 1 | ||
| 2102 | like | 1 | 0 | 0 | 1 | ||
| 2103 | no_response | 1 | 0 | 0 | 1 | ||
| 2110 | dislike | 3 | 0 | 0 | 3 | ||
| 2200 | like | 22 | 2 | 0 | 24 | ||
| 2210 | dislike | 11 | 0 | 0 | 11 | ||
| All | 304 | 4 | 1 | 309 |
is_anchor: False
(50, 33)
| log_flags | 0 | All | |||
|---|---|---|---|---|---|
| data_group | ccode | study_bin_id | study_response | ||
| sub000 | 2 | 2100 | like | 9 | 9 |
| 2101 | like | 1 | 1 | ||
| 2102 | like | 1 | 1 | ||
| 2103 | no_response | 1 | 1 | ||
| 2110 | dislike | 3 | 3 | ||
| 2200 | like | 24 | 24 | ||
| 2210 | dislike | 11 | 11 | ||
| All | 50 | 50 |
test phase codemap
define the template
plug in the individual items
save the result
[9]:
# test phase codemap name
pm_test_codemap_f = MDE_HOME / "mkpy/pm_test_codemap.tsv"
# test phase codemap column names
pm_test_codemap_cols = [
"regexp", "test_bin_id", "animacy", "stimulus", "test_response", "accuracy"
] + list(pm_items.columns)
pm_test_codemap = pd.read_csv(pm_test_codemap_f, sep="\t")
display(pm_test_codemap.shape)
display(pm_test_codemap)
(706, 12)
| regexp | test_bin_id | animacy | stimulus | test_response | accuracy | ccode | phase | condition_id | item_id | jpg | scn | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | (#[1234]) | 0 | cal | cal | cal | cal | 0 | test | cal | -1 | cal | cal |
| 1 | (#[1234]) | 10 | _any | _any | _any | _any | 1 | test | _any | -1 | _any | _any |
| 2 | (#[1]) | 11 | animate | distractor | _any | _any | 1 | test | 1 | -1 | _any | _any |
| 3 | (#[2]) | 12 | inanimate | distractor | _any | _any | 1 | test | 2 | -1 | _any | _any |
| 4 | (#[3]) | 13 | animate | studied | _any | _any | 1 | test | 3 | -1 | _any | _any |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 701 | (#2) 2064 8 (105) | 1202 | inanimate | distractor | new | cr | 1 | test | 2 | 105 | barn | testp1 |
| 702 | (#2) 8 (105) 1040 | 1210 | inanimate | distractor | old | fa | 1 | test | 2 | 105 | barn | testp1 |
| 703 | (#2) 8 1040 (105) | 1211 | inanimate | distractor | old | fa | 1 | test | 2 | 105 | barn | testp1 |
| 704 | (#2) 1040 8 (105) | 1212 | inanimate | distractor | old | fa | 1 | test | 2 | 105 | barn | testp1 |
| 705 | (#2) 8 (105) (?!(2064|1040)) | 1203 | inanimate | distractor | none | nr | 1 | test | 2 | 105 | barn | testp1 |
706 rows × 12 columns
test phase get_event_table(codemap)
[10]:
pm_test_event_table = pm_h5.get_event_table(pm_test_codemap_f)
searching codes in: sub000/dblock_0
/mnt/cube/home/turbach/TPU_Projects/mkpy/mkpy/mkh5.py:1059: UserWarning:
As of mkpy 0.2.0 to match events with a codemap regexp pattern, the
ccode column in pm_test_codemap.tsv must also match the log_ccode
in the datablock. If this behavior is not desired, delete or rename
the ccode column in the codemap.
warnings.warn(msg)
searching codes in: sub000/dblock_1
searching codes in: sub000/dblock_2
searching codes in: sub000/dblock_3
inspect test phase event table
[11]:
print("test phase (shape):", pm_test_event_table.shape)
print("test phase columns:", pm_test_event_table.columns)
# select some columns to show
example_columns = [
"dblock_path", "dblock_ticks", "log_evcodes", "log_ccodes", "log_flags",
"regexp", "match_code",
"phase", "test_bin_id", "test_response", "accuracy",
]
# first few stimulus events, coded for response
display(pm_test_event_table[example_columns].query("test_bin_id > 1000").head())
# last few calibration pulse events
display(pm_test_event_table[example_columns].tail())
for is_anchor in [True, False]:
print("is_anchor: ", is_anchor)
events = pm_test_event_table.query("is_anchor == @is_anchor ")
display(
pd.crosstab(
[
events.data_group,
# events.ccode,
events.test_bin_id,
events.animacy,
events.stimulus,
events.test_response
],
[
events.accuracy
],
margins=True
)
)
test phase (shape): (609, 35)
test phase columns: Index(['data_group', 'dblock_path', 'dblock_tick_idx', 'dblock_ticks',
'crw_ticks', 'raw_evcodes', 'log_evcodes', 'log_ccodes', 'log_flags',
'epoch_match_tick_delta', 'epoch_ticks', 'dblock_srate', 'match_group',
'idx', 'dlim', 'anchor_str', 'match_str', 'anchor_code', 'match_code',
'anchor_tick', 'match_tick', 'anchor_tick_delta', 'is_anchor', 'regexp',
'test_bin_id', 'animacy', 'stimulus', 'test_response', 'accuracy',
'ccode', 'phase', 'condition_id', 'item_id', 'jpg', 'scn'],
dtype='object')
| dblock_path | dblock_ticks | log_evcodes | log_ccodes | log_flags | regexp | match_code | phase | test_bin_id | test_response | accuracy | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 204 | sub000/dblock_1 | 611 | 1 | 1 | 0 | (#1) 2064 8 (127) | 1 | test | 1102 | new | cr |
| 205 | sub000/dblock_1 | 1061 | 127 | 1 | 0 | (#1) 2064 8 (127) | 127 | test | 1102 | new | cr |
| 206 | sub000/dblock_1 | 2062 | 4 | 1 | 0 | (#4) 8 (129) 1040 | 4 | test | 1400 | old | hit |
| 207 | sub000/dblock_1 | 2516 | 129 | 1 | 0 | (#4) 8 (129) 1040 | 129 | test | 1400 | old | hit |
| 208 | sub000/dblock_1 | 3767 | 2 | 1 | 0 | (#2) 8 (185) 2064 | 2 | test | 1200 | new | cr |
| dblock_path | dblock_ticks | log_evcodes | log_ccodes | log_flags | regexp | match_code | phase | test_bin_id | test_response | accuracy | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 854 | sub000/dblock_3 | 27315 | 4 | 0 | 0 | (#[1234]) | 4 | test | 0 | cal | cal |
| 855 | sub000/dblock_3 | 27444 | 2 | 0 | 0 | (#[1234]) | 2 | test | 0 | cal | cal |
| 856 | sub000/dblock_3 | 27573 | 3 | 0 | 0 | (#[1234]) | 3 | test | 0 | cal | cal |
| 857 | sub000/dblock_3 | 27703 | 4 | 0 | 0 | (#[1234]) | 4 | test | 0 | cal | cal |
| 858 | sub000/dblock_3 | 27832 | 2 | 0 | 0 | (#[1234]) | 2 | test | 0 | cal | cal |
is_anchor: True
| accuracy | _any | cal | cr | fa | hit | All | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| data_group | test_bin_id | animacy | stimulus | test_response | ||||||
| sub000 | 0 | cal | cal | cal | 0 | 209 | 0 | 0 | 0 | 209 |
| 10 | _any | _any | _any | 100 | 0 | 0 | 0 | 0 | 100 | |
| 11 | animate | distractor | _any | 15 | 0 | 0 | 0 | 0 | 15 | |
| 12 | inanimate | distractor | _any | 35 | 0 | 0 | 0 | 0 | 35 | |
| 13 | animate | studied | _any | 15 | 0 | 0 | 0 | 0 | 15 | |
| 14 | inanimate | studied | _any | 35 | 0 | 0 | 0 | 0 | 35 | |
| 1100 | animate | distractor | new | 0 | 0 | 14 | 0 | 0 | 14 | |
| 1102 | animate | distractor | new | 0 | 0 | 1 | 0 | 0 | 1 | |
| 1200 | inanimate | distractor | new | 0 | 0 | 32 | 0 | 0 | 32 | |
| 1201 | inanimate | distractor | new | 0 | 0 | 1 | 0 | 0 | 1 | |
| 1202 | inanimate | distractor | new | 0 | 0 | 1 | 0 | 0 | 1 | |
| 1210 | inanimate | distractor | old | 0 | 0 | 0 | 1 | 0 | 1 | |
| 1300 | animate | studied | old | 0 | 0 | 0 | 0 | 15 | 15 | |
| 1400 | inanimate | studied | old | 0 | 0 | 0 | 0 | 35 | 35 | |
| All | 200 | 209 | 49 | 1 | 50 | 509 |
is_anchor: False
| accuracy | cr | fa | hit | All | ||||
|---|---|---|---|---|---|---|---|---|
| data_group | test_bin_id | animacy | stimulus | test_response | ||||
| sub000 | 1100 | animate | distractor | new | 14 | 0 | 0 | 14 |
| 1102 | animate | distractor | new | 1 | 0 | 0 | 1 | |
| 1200 | inanimate | distractor | new | 32 | 0 | 0 | 32 | |
| 1201 | inanimate | distractor | new | 1 | 0 | 0 | 1 | |
| 1202 | inanimate | distractor | new | 1 | 0 | 0 | 1 | |
| 1210 | inanimate | distractor | old | 0 | 1 | 0 | 1 | |
| 1300 | animate | studied | old | 0 | 0 | 15 | 15 | |
| 1400 | inanimate | studied | old | 0 | 0 | 35 | 35 | |
| All | 49 | 1 | 50 | 100 |
Prune study phase event table for epochs and tag with test phase responses
The study events were double counted as stim only and again as stim + response in order to verify code mapping
Now drop redundant stim-only events
[12]:
# ----------------------------------------------
# prune event tables to unique single trials
# ----------------------------------------------
# study phase response tagged single trials are coded with study table bin id > 2000
pm_study_events_for_epochs = pm_study_event_table.query(
"is_anchor==True and study_bin_id >= 2000"
).copy().set_index("item_id").sort_index()
# test phase response-tagged single trials are coded with test table bin id > 1000
pm_test_events_for_epochs = pm_test_event_table.query(
"is_anchor==True and test_bin_id >= 1000"
).copy().set_index("item_id").sort_index()
[13]:
# --------------------------------------------------------
# update study phase events with test phase responses
# --------------------------------------------------------
# just for summary display ...
display_cols = ["match_code", "anchor_code", "log_evcodes", "phase", "study_response"]
# test response tags
test_s_r_cols = ["test_response", "accuracy"]
print("Study phase items before ...")
display(pm_study_events_for_epochs.shape)
display(pm_study_events_for_epochs[display_cols].head())
# align the test phase subsequent responses with the study phase items
pm_study_events_for_epochs = (
pm_study_events_for_epochs
.join(
pm_test_events_for_epochs[test_s_r_cols],
how="left",
on="item_id",
)
)
print("Study phase items after joining test phase responses ...")
display(pm_study_events_for_epochs.shape)
display(pm_study_events_for_epochs[display_cols + test_s_r_cols].head())
Study phase items before ...
(50, 32)
| match_code | anchor_code | log_evcodes | phase | study_response | |
|---|---|---|---|---|---|
| item_id | |||||
| 101 | 1 | 1 | 1 | study | dislike |
| 103 | 2 | 2 | 2 | study | dislike |
| 106 | 1 | 1 | 1 | study | like |
| 107 | 2 | 2 | 2 | study | like |
| 112 | 2 | 2 | 2 | study | dislike |
Study phase items after joining test phase responses ...
(50, 34)
| match_code | anchor_code | log_evcodes | phase | study_response | test_response | accuracy | |
|---|---|---|---|---|---|---|---|
| item_id | |||||||
| 101 | 1 | 1 | 1 | study | dislike | old | hit |
| 103 | 2 | 2 | 2 | study | dislike | old | hit |
| 106 | 1 | 1 | 1 | study | like | old | hit |
| 107 | 2 | 2 | 2 | study | like | old | hit |
| 112 | 2 | 2 | 2 | study | dislike | old | hit |
[14]:
# --------------------------------------------------------
# update test phase event table with study phase responses
# ---------------------------------------------------------
# just for summary display ...
display_cols = ["match_code", "anchor_code", "log_evcodes", "phase"]
# study response tags to map to the other phase
study_s_r_cols = ["study_response"]
print("Test phase items before:", pm_test_events_for_epochs.shape)
display(pm_test_events_for_epochs[display_cols].head(12))
# align the study phase like/dislike responses with the test phase items
pm_test_events_for_epochs = (
pm_test_events_for_epochs
.join(
pm_study_events_for_epochs["study_response"],
how="left",
on="item_id",
)
)
print("Test phase items after", pm_test_events_for_epochs.shape)
display(pm_test_events_for_epochs[display_cols + study_s_r_cols].head(12))
Test phase items before: (100, 34)
| match_code | anchor_code | log_evcodes | phase | |
|---|---|---|---|---|
| item_id | ||||
| 101 | 3 | 3 | 3 | test |
| 102 | 1 | 1 | 1 | test |
| 103 | 4 | 4 | 4 | test |
| 104 | 2 | 2 | 2 | test |
| 105 | 2 | 2 | 2 | test |
| 106 | 3 | 3 | 3 | test |
| 107 | 4 | 4 | 4 | test |
| 108 | 1 | 1 | 1 | test |
| 109 | 2 | 2 | 2 | test |
| 110 | 1 | 1 | 1 | test |
| 111 | 2 | 2 | 2 | test |
| 112 | 4 | 4 | 4 | test |
Test phase items after (100, 35)
| match_code | anchor_code | log_evcodes | phase | study_response | |
|---|---|---|---|---|---|
| item_id | |||||
| 101 | 3 | 3 | 3 | test | dislike |
| 102 | 1 | 1 | 1 | test | NaN |
| 103 | 4 | 4 | 4 | test | dislike |
| 104 | 2 | 2 | 2 | test | NaN |
| 105 | 2 | 2 | 2 | test | NaN |
| 106 | 3 | 3 | 3 | test | like |
| 107 | 4 | 4 | 4 | test | like |
| 108 | 1 | 1 | 1 | test | NaN |
| 109 | 2 | 2 | 2 | test | NaN |
| 110 | 1 | 1 | 1 | test | NaN |
| 111 | 2 | 2 | 2 | test | NaN |
| 112 | 4 | 4 | 4 | test | dislike |
study phase set_epochs(name, pre, post)
[15]:
pm_h5.set_epochs("study_ms1500", pm_study_events_for_epochs, -750, 750)
Sanitizing event table data types for mkh5 epochs table ...
study phase export_epochs(name)
[16]:
pm_study_epochs_f = DOCS_DATA / "sub000pm.study_ms1500.epochs.feather"
pm_h5.export_epochs("study_ms1500", pm_study_epochs_f, file_format="feather")
test phase set_epochs(name, pre, post)
[17]:
pm_h5.set_epochs("test_ms1500", pm_test_events_for_epochs, -750, 750)
Sanitizing event table data types for mkh5 epochs table ...
test phase export_epochs(name)
[18]:
pm_test_epochs_f = DOCS_DATA / "sub000pm.test_ms1500.epochs.feather"
pm_h5.export_epochs("test_ms1500", pm_test_epochs_f, file_format="feather")
Analyze the epochs
Is there a difference between like and dislike at study?
Is there a difference between old and new at test?
If so is it the same for liked and disliked?
[19]:
# matplotlib line colors, background, fonts
plt.style.use("bmh")
Study phase time-domain average ERPs
[20]:
# load and sanitize epoch_id for pandas index
pm_study_epochs = pd.read_feather(pm_study_epochs_f)
pm_study_epochs['epoch_id'] = pm_study_epochs['epoch_id'].astype('int')
# exclude epochs flagged for artifacts
pm_study_epochs = epf.drop_bad_epochs(
pm_study_epochs,
bads_column="log_flags",
epoch_id="epoch_id",
time="match_time"
)
# check the good event counts after dropping artifacts
pm_study_good_events = pm_study_epochs.query("match_time == 0")
print("After excluding EEG artifacts")
display(
pd.crosstab(
[
pm_study_good_events.ccode,
pm_study_good_events.animacy,
pm_study_good_events.study_response,
],
[
pm_study_good_events.log_flags
],
margins=True
)
)
# for illustration ...
midline = ["MiPf", "MiCe", "MiPa", "MiOc"]
# select COLUMNS: epoch index, timestamps, event tags, and midline EEG columns
midline_epochs = pm_study_epochs[["epoch_id", "match_time", "animacy", "study_response"] + midline]
# select ROWS: use tag values to pick and choose
midline_epochs = midline_epochs.query("study_response in ['like', 'dislike']")
# center each channel
midline_epochs = epf.center_eeg(
midline_epochs,
midline,
-750, 0,
epoch_id="epoch_id",
time="match_time"
)
After excluding EEG artifacts
| log_flags | 0 | All | ||
|---|---|---|---|---|
| ccode | animacy | study_response | ||
| 2 | animate | dislike | 3 | 3 |
| like | 11 | 11 | ||
| no_response | 1 | 1 | ||
| inanimate | dislike | 11 | 11 | |
| like | 22 | 22 | ||
| All | 48 | 48 |
[21]:
for grp in [["animacy"], ["study_response"], ["animacy", "study_response"]]:
# compute domain average by stim type
midline_erps = midline_epochs.groupby(
grp + ["match_time"]
).mean().reset_index()
# plot
f, axs = plt.subplots(1, 4, figsize=(14,8), sharex=True)
for rep, erp in midline_erps.groupby(grp):
for axi, chan in enumerate(midline):
# mark onset
axs[axi].axhline(0, color='gray')
# plot erp
axs[axi].plot(
erp[chan],
erp["match_time"],
label=f"{rep}",
lw=2,
)
# channel
axs[axi].set(xlim=(-15, 20), xlabel=chan)
axs[0].legend(loc="lower right")
axs[0].set_title(f"Study phase:\n{':'.join(grp)}", fontsize=12, loc="left")
f.tight_layout()
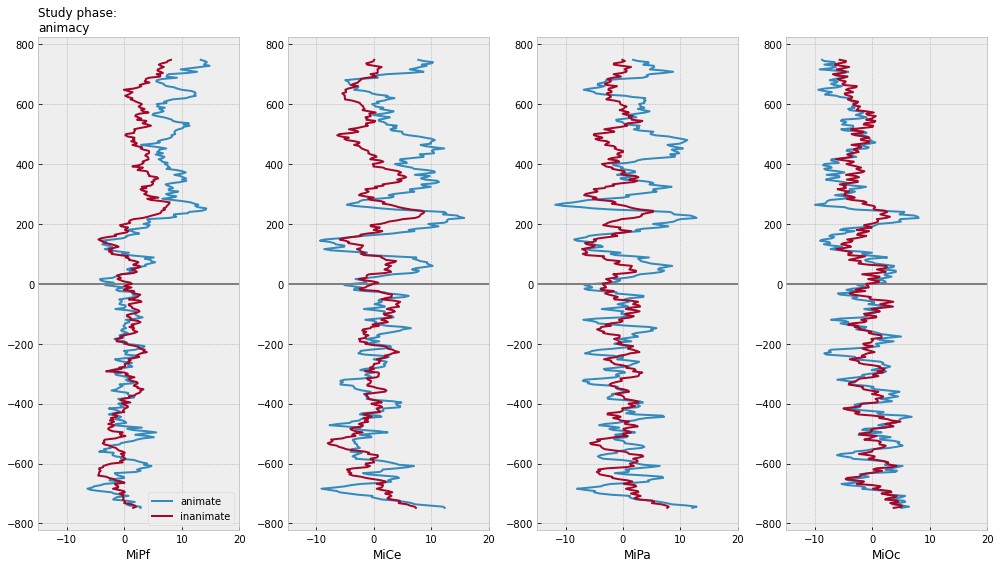
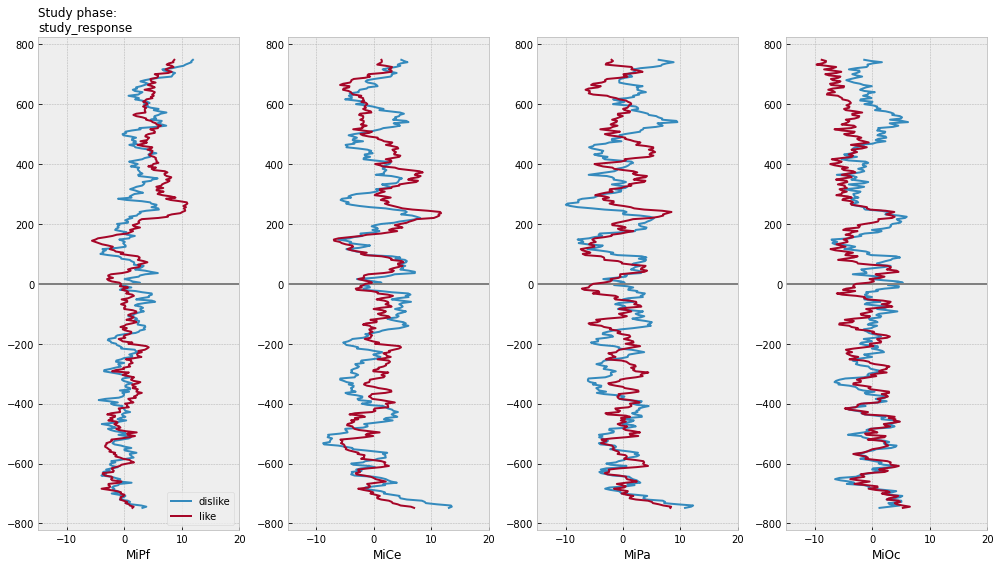
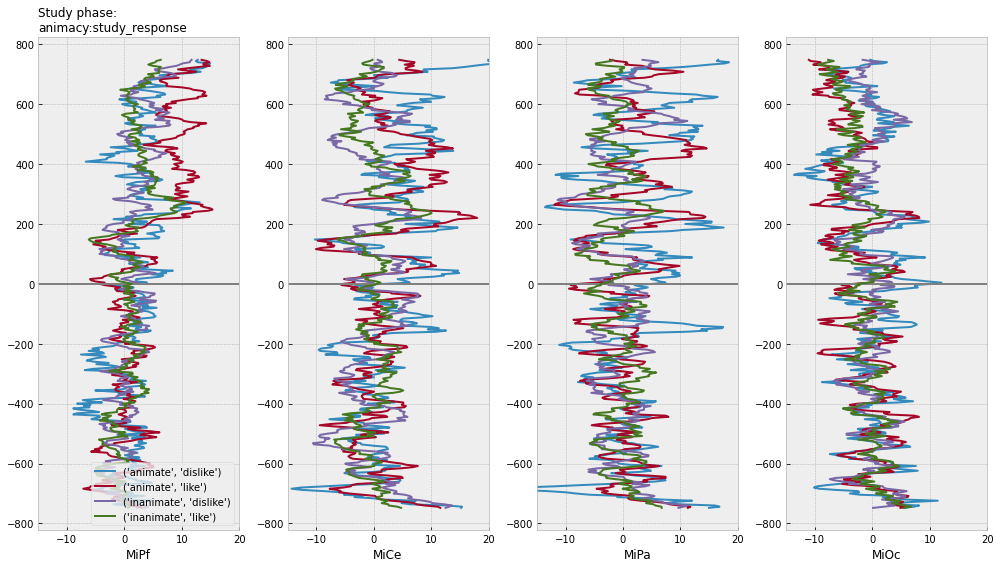
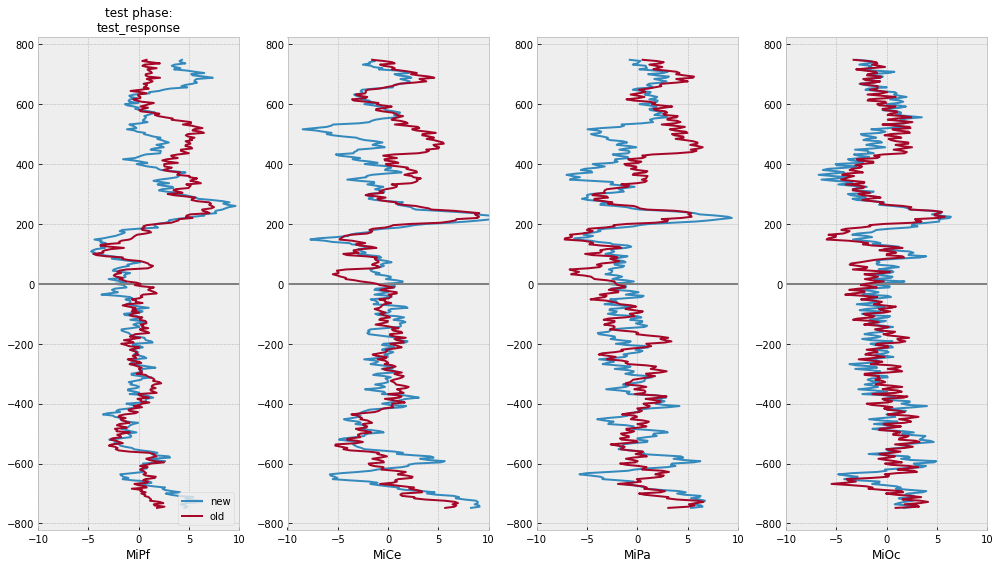
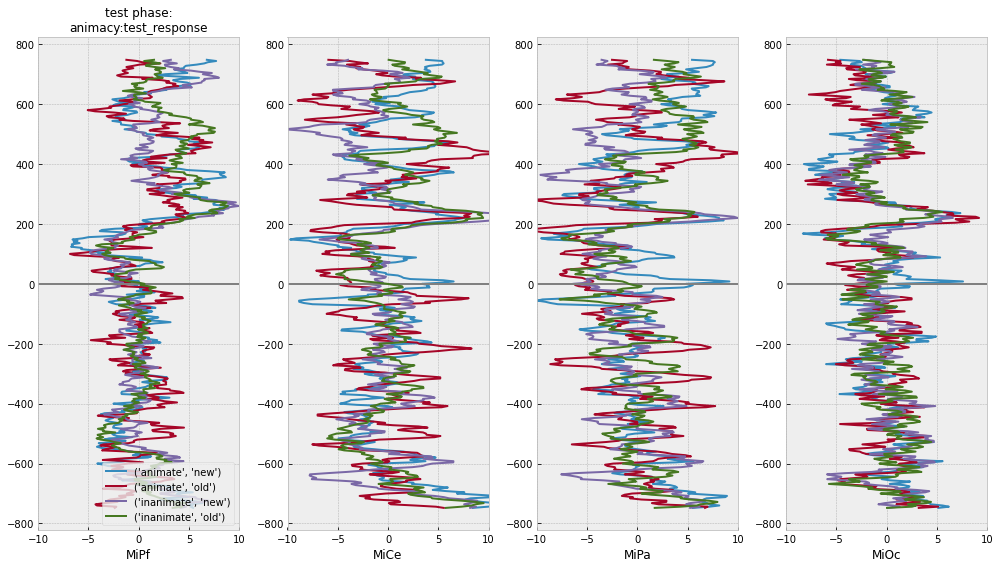
test phase time-domain average ERPs
[22]:
# load and sanitize epoch_id for pandas index
pm_test_epochs = pd.read_feather(pm_test_epochs_f)
pm_test_epochs['epoch_id'] = pm_test_epochs['epoch_id'].astype('int')
print(pm_test_epochs.columns)
# drop EEG epochs tagged as bad
pm_test_epochs = epf.drop_bad_epochs(
pm_test_epochs,
bads_column="log_flags",
epoch_id="epoch_id",
time="match_time"
)
# check the good event counts after dropping artifacts
pm_test_good_events = pm_test_epochs.query("match_time == 0")
print("After excluding EEG artifacts")
display(
pd.crosstab(
[
pm_test_good_events.ccode,
pm_test_good_events.animacy,
pm_test_good_events.test_response,
],
[
pm_test_good_events.log_flags
],
margins=True
)
)
Index(['epoch_id', 'data_group', 'dblock_path', 'dblock_tick_idx',
'dblock_ticks', 'crw_ticks', 'raw_evcodes', 'log_evcodes', 'log_ccodes',
'log_flags', 'epoch_match_tick_delta', 'epoch_ticks', 'dblock_srate',
'match_group', 'idx', 'dlim', 'anchor_str', 'match_str', 'anchor_code',
'match_code', 'anchor_tick', 'match_tick', 'anchor_tick_delta',
'is_anchor', 'regexp', 'test_bin_id', 'animacy', 'stimulus',
'test_response', 'accuracy', 'ccode', 'phase', 'condition_id', 'jpg',
'scn', 'study_response', 'match_time', 'anchor_time',
'anchor_time_delta', 'diti_t_0', 'diti_hop', 'diti_len', 'pygarv',
'lle', 'lhz', 'MiPf', 'LLPf', 'RLPf', 'LMPf', 'RMPf', 'LDFr', 'RDFr',
'LLFr', 'RLFr', 'LMFr', 'RMFr', 'LMCe', 'RMCe', 'MiCe', 'MiPa', 'LDCe',
'RDCe', 'LDPa', 'RDPa', 'LMOc', 'RMOc', 'LLTe', 'RLTe', 'LLOc', 'RLOc',
'MiOc', 'A2', 'HEOG', 'rle', 'rhz'],
dtype='object')
After excluding EEG artifacts
| log_flags | 0 | All | ||
|---|---|---|---|---|
| ccode | animacy | test_response | ||
| 1 | animate | new | 14 | 14 |
| old | 13 | 13 | ||
| inanimate | new | 31 | 31 | |
| old | 35 | 35 | ||
| All | 93 | 93 |
[23]:
# for illustration ...
midline = ["MiPf", "MiCe", "MiPa", "MiOc"]
# select COLUMNS: epoch index, timestamps, event tags, and midline EEG columns
midline_epochs = pm_test_epochs[["epoch_id", "match_time", "animacy", "test_response"] + midline]
# select ROWS: use tag values to pick and choose
midline_epochs = midline_epochs.query("test_response in ['old', 'new']")
# center each channel
midline_epochs = epf.center_eeg(
midline_epochs,
midline,
-750, 0,
epoch_id="epoch_id",
time="match_time"
)
[24]:
# plot separately and together
for grp in [["animacy"], ["test_response"], ["animacy", "test_response"]]:
# compute domain average by stim type
midline_erps = midline_epochs.groupby(
grp + ["match_time"]
).mean().reset_index()
# plot
f, axs = plt.subplots(1, 4, figsize=(14,8))
for (rep), erp in midline_erps.groupby(grp):
for axi, chan in enumerate(midline):
# mark onset
axs[axi].axhline(0, color='gray')
# plot erp
axs[axi].plot(
erp[chan],
erp["match_time"],
label=f"{rep}",
lw=2,
)
axs[axi].set(xlim=(-10, 10), xlabel=chan)
axs[0].legend(loc="lower right")
axs[0].set_title(f"test phase:\n{':'.join(grp)}", fontsize=12)
f.tight_layout()
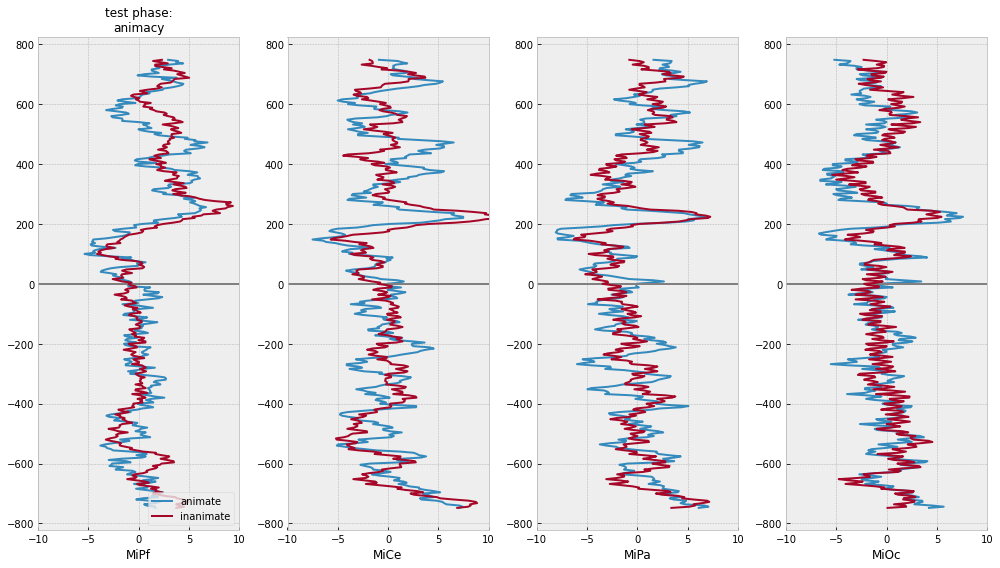
Appendix¶
Python code to generate the study and test code maps from the stimulus files
[25]:
%%bash
cat ${MDE_HOME}/scripts/make_pm_codemaps.py
"""demonstrate programmatic codemap generation"""
import re
import pandas as pd
from make_zenodo_files import MDE_HOME
# stim info scraped from .scn scenario files
PM_ITEM_ID_BY_SCN_F = MDE_HOME / "mkpy/pm_item_id_by_scn.tsv"
# study phase codemap
PM_STUDY_CODEMAP_F = MDE_HOME / "mkpy/pm_study_codemap.tsv"
# test phase codemap
PM_TEST_CODEMAP_F = MDE_HOME / "mkpy/pm_test_codemap.tsv"
def make_pm_item_id_by_scn():
"""scrape stimuli out of the presentation files and extract code information"""
stim_split_re = re.compile(
r"(?P<soa>\d+)\s+(?P<dur>\d+)\s+(?P<evcode>\d+)\s+"
r"(?P<type>....)=(?P<stim>.+)[\.]"
)
scn_paths = sorted(MDE_HOME.glob("mkstim/pictmem/*.scn"))
header_str = "".join(
[
f"{col:>16}"
for col in ["ccode", "phase", "condition_id", "item_id", "jpg", "scn"]
]
)
items = [header_str]
for scn_path in scn_paths:
with open(scn_path, "r") as _fh:
scn = scn_path.stem
phase = None
ccode = None
for phase, ccode in [("study", 2), ("test", 1)]:
if phase in str(scn_path):
break
assert phase is not None and ccode is not None
for line in _fh.readlines():
fields = stim_split_re.match(line)
if fields is None:
continue
evcode = int(fields["evcode"])
if evcode > 0 and evcode <= 4:
assert fields["type"] == "jpeg"
stim = fields["stim"]
cond = evcode
if evcode > 100:
item = evcode
items.append(
"".join(
[
f"{val:>16}"
for val in [ccode, phase, cond, item, stim, scn]
]
)
)
with open(PM_ITEM_ID_BY_SCN_F, "w") as _fh:
_fh.write("\n".join(items))
_fh.write("\n")
def make_pm_study_phase_codemap():
""" Study phase """
# read the item information table
pm_items = pd.read_csv(PM_ITEM_ID_BY_SCN_F, delim_whitespace=True).query(
"scn in ['studyp1', 'testp1']"
)
# code map
pm_study_codemap_cols = [
"regexp",
"study_bin_id",
"animacy",
"study_response",
] + list(pm_items.columns)
# stimulus-response tag template as a Python dictionary
# The key:val pair says "this code sequence gets these tags"
# The ITEM_ID string will be replaced by the actual 3-digit item number
study_code_tags = {
"(#[12]) 8 (ITEM_ID) 1040": (2000, "like"),
"(#[12]) 8 1040 (ITEM_ID)": (2001, "like"),
"(#[12]) 1040 8 (ITEM_ID)": (2002, "like"),
"(#[12]) 8 (ITEM_ID) 2064": (2100, "dislike"),
"(#[12]) 8 2064 (ITEM_ID)": (2101, "dislike"),
"(#[12]) 2064 8 (ITEM_ID)": (2102, "dislike"),
"(#[12]) 8 (ITEM_ID) (?!(1040|2064))": (2003, "no_response"),
}
# the new 4-digit "study_bin_id" tag re-codes the match event 1 or
# 2 with more information
#
# phase animacy response response_timing
# phase: study=2
# animacy: 1=animate, 2=inanimate
# response(0=like, 1=dislike)
# response timing: 0=prompted,1,2 anticipation, 3=no response)
#
study_code_tags = {
# animate
"(#[1]) 8 (ITEM_ID) 1040": (2100, "animate", "like"),
"(#[1]) 8 1040 (ITEM_ID)": (2101, "animate", "like"),
"(#[1]) 1040 8 (ITEM_ID)": (2102, "animate", "like"),
"(#[1]) 8 (ITEM_ID) 2064": (2110, "animate", "dislike"),
"(#[1]) 8 2064 (ITEM_ID)": (2111, "animate", "dislike"),
"(#[1]) 2064 8 (ITEM_ID)": (2112, "animate", "dislike"),
"(#[1]) 8 (ITEM_ID) (?!(1040|2064))": (2103, "animate", "no_response"),
# inanimate
"(#[2]) 8 (ITEM_ID) 1040": (2200, "inanimate", "like"),
"(#[2]) 8 1040 (ITEM_ID)": (2201, "inanimate", "like"),
"(#[2]) 1040 8 (ITEM_ID)": (2202, "inanimate", "like"),
"(#[2]) 8 (ITEM_ID) 2064": (2210, "inanimate", "dislike"),
"(#[2]) 8 2064 (ITEM_ID)": (2211, "inanimate", "dislike"),
"(#[2]) 2064 8 (ITEM_ID)": (2212, "inanimate", "dislike"),
"(#[2]) 8 (ITEM_ID) (?!(1040|2064))": (2203, "inanimate", "no_response"),
}
#
# Build a list of codemap lines.
# The first line says *any* code matching 1 or 2 gets the tags 200, "_any", 2, ... etc.
# This tags all matching stimulus events, it is not contingent the response.
# It is not necessary but it is useful here, we will see why shortly.
study_code_map = [
("(#[1234])", 0, "cal", "cal", 0, "study", 0, -1, "cal", "cal"),
("(#[12])", 200, "_any", "_any", 2, "study", 2, -1, "_any", "_any"),
]
# plug each row of the pictmem item info into the template and append the
# result to the list of codemap lines
for idx, row in pm_items.query("phase == 'study'").iterrows():
for pattern, tags in study_code_tags.items():
code_tags = (
pattern.replace(
"ITEM_ID", str(row.item_id)
), # current item number goes in the template
*(str(t) for t in tags),
*(str(c) for c in row), # this adds the rest of the item to the tags
)
study_code_map.append(code_tags)
# convert the list of lines to a pandas.DataFrame and save as a tab separated text file
pm_study_codemap = pd.DataFrame(study_code_map, columns=pm_study_codemap_cols)
pm_study_codemap.to_csv(PM_STUDY_CODEMAP_F, sep="\t", index=False)
print(pm_study_codemap.shape)
print(pm_study_codemap)
def make_pm_test_phase_codemap():
# read the item information table
pm_items = pd.read_csv(PM_ITEM_ID_BY_SCN_F, delim_whitespace=True).query(
"scn in ['studyp1', 'testp1']"
)
# test phase codemap column names
pm_test_codemap_cols = [
"regexp",
"test_bin_id",
"animacy",
"stimulus",
"test_response",
"accuracy",
] + list(pm_items.columns)
# test phase template: stimulus, old/new response (include pre-prompt anticipations)
test_code_tags = {
# new stim animate
"(#1) 8 (ITEM_ID) 2064": (1100, "animate", "distractor", "new", "cr"),
"(#1) 8 2064 (ITEM_ID)": (1101, "animate", "distractor", "new", "cr"),
"(#1) 2064 8 (ITEM_ID)": (1102, "animate", "distractor", "new", "cr"),
"(#1) 8 (ITEM_ID) 1040": (1110, "animate", "distractor", "old", "fa"),
"(#1) 8 1040 (ITEM_ID)": (1111, "animate", "distractor", "old", "fa"),
"(#1) 1040 8 (ITEM_ID)": (1112, "animate", "distractor", "old", "fa"),
"(#1) 8 (ITEM_ID) (?!(2064|1040))": (
1103,
"animate",
"distractor",
"none",
"nr",
),
# new stim inanimate
"(#2) 8 (ITEM_ID) 2064": (1200, "inanimate", "distractor", "new", "cr"),
"(#2) 8 2064 (ITEM_ID)": (1201, "inanimate", "distractor", "new", "cr"),
"(#2) 2064 8 (ITEM_ID)": (1202, "inanimate", "distractor", "new", "cr"),
"(#2) 8 (ITEM_ID) 1040": (1210, "inanimate", "distractor", "old", "fa"),
"(#2) 8 1040 (ITEM_ID)": (1211, "inanimate", "distractor", "old", "fa"),
"(#2) 1040 8 (ITEM_ID)": (1212, "inanimate", "distractor", "old", "fa"),
"(#2) 8 (ITEM_ID) (?!(2064|1040))": (
1203,
"inanimate",
"distractor",
"none",
"nr",
),
# old stim animate
"(#3) 8 (ITEM_ID) 1040": (1300, "animate", "studied", "old", "hit"),
"(#3) 8 1040 (ITEM_ID)": (1301, "animate", "studied", "old", "hit"),
"(#3) 1040 8 (ITEM_ID)": (1302, "animate", "studied", "old", "hit"),
"(#3) 8 (ITEM_ID) 2064": (1310, "animate", "studied", "new", "miss"),
"(#3) 8 2064 (ITEM_ID)": (1311, "animate", "studied", "new", "miss"),
"(#3) 2064 8 (ITEM_ID)": (1312, "animate", "studied", "new", "miss"),
"(#3) 8 (ITEM_ID) (?!(2064|1040))": (1303, "animate", "studied", "none", "nr"),
# old stim inanimate
"(#4) 8 (ITEM_ID) 1040": (1400, "inanimate", "studied", "old", "hit"),
"(#4) 8 1040 (ITEM_ID)": (1401, "inanimate", "studied", "old", "hit"),
"(#4) 1040 8 (ITEM_ID)": (1402, "inanimate", "studied", "old", "hit"),
"(#4) 8 (ITEM_ID) 2064": (1410, "inanimate", "studied", "new", "miss"),
"(#4) 8 2064 (ITEM_ID)": (1411, "inanimate", "studied", "new", "miss"),
"(#4) 2064 8 (ITEM_ID)": (1412, "inanimate", "studied", "new", "miss"),
"(#4) 8 (ITEM_ID) (?!(2064|1040))": (
1403,
"inanimate",
"studied",
"none",
"nr",
),
}
# initialize the code map to tag stimulus codes, not response contingent
test_code_map = [
(
"(#[1234])",
0,
"cal",
"cal",
"cal",
"cal",
0,
"test",
"cal",
"-1",
"cal",
"cal",
),
(
"(#[1234])",
10,
"_any",
"_any",
"_any",
"_any",
1,
"test",
"_any",
"-1",
"_any",
"_any",
),
(
"(#[1])",
11,
"animate",
"distractor",
"_any",
"_any",
1,
"test",
1,
"-1",
"_any",
"_any",
),
(
"(#[2])",
12,
"inanimate",
"distractor",
"_any",
"_any",
1,
"test",
2,
"-1",
"_any",
"_any",
),
(
"(#[3])",
13,
"animate",
"studied",
"_any",
"_any",
1,
"test",
3,
"-1",
"_any",
"_any",
),
(
"(#[4])",
14,
"inanimate",
"studied",
"_any",
"_any",
1,
"test",
4,
"-1",
"_any",
"_any",
),
]
# iterate through the item info and plug the item number into the template lines
for idx, row in pm_items.query("phase == 'test'").iterrows():
for pattern, tags in test_code_tags.items():
# condition_id is 1, 2, 3, or 4 only plug into the relevant template lines.
if re.match(r"^\(#" + str(row.condition_id), pattern):
code_tags = (
pattern.replace("ITEM_ID", str(row.item_id)),
tags[0],
*(str(t) for t in tags[1:]),
*(str(c) for c in row),
)
test_code_map.append(code_tags)
pm_test_codemap = pd.DataFrame(test_code_map, columns=pm_test_codemap_cols)
# write test demo phase codemap
pm_test_codemap.to_csv(PM_TEST_CODEMAP_F, sep="\t", index=False)
print(pm_test_codemap.shape)
print(pm_test_codemap)
if __name__ == "__main__":
make_pm_item_id_by_scn()
make_pm_study_phase_codemap()
make_pm_test_phase_codemap()