Background: EEG, events, and epochs¶
event codes vs. variables¶
In brainwave experiments, the timing of events that occur while the EEG data are recorded is often tracked by logging a time-stamped integer event code. The timestamp indicates when the event occured in relation to the EEG recording and the integer value identifies the type of event. For example, the event code may indicate the stimulus (1 = beep, 2 = boop) or response (3 for a left hand button press, 4 for a right).
In very simple single factor experiments, the integer code itself can serve as a predictor variable for modeling the data. For instance, integer event codes for two stimulus types, beep = 0 and boop = 1 treatment code the stimulus variable with beep as the reference level. Alternatively the variable can be sum (deviation) coded as beep = -1 and boop = 1. So in this kind of case, integer event codes can be read off the data log and dropped into a model (modeling software) without modification. For the vast majority of real science, event codes must be re-coded, i.e., systematically transformed or mapped, to other representations for purposes of modeling the data. There are several reasons.
The event codes logged with the data recording are fossilized – encased in digital amber, immutable. They can encode a treatment coding or a sum coding but not both. If a different coding is required (Reviewer #2 insists), the original codes cannot be used directly. This is not a problem in practice because you can confess to your modeling software that the integer codes are really just category labels anyway and then specify which categorical coding scheme to use for constructing the model (design) matrix. It is convenient that for this simple special case, someone else has solved the problem of re-mapping our integer event codes for purposes of modeling the data and we can get on with our work. The important point is that even in this simplest of cases, the event codes as originally logged with the data had to be recoded after the fact for modeling the data. Recoding fossilized event codes is the general problem, and statistical modeling sofware does not have a general solution.
The core issue is that models need quantities and although event codes are logged as integers, they are actually simple event labels or tags that uniquely identify the event (type or token) that occurred. In place of codes 1 and 2, labels like “a” and “b” or “beep” and “boop” would do just as well if the data acquisition hardware were able to log and timestamp character strings. Event codes are simple in the sense that contrasts with complex: a unit with no smaller parts. To uniquely code a set of four events, the following codings do exactly the same job: {1, 2, 3, 4}, {“a”, “b”, “c”, “d”}, {11, 12, 21, 22}. To simplify computation, the current crop of statistical modeling software (R, statsmodels, SAS, SPSS, SYSTAT) wants the data arranged in a tabular format: observations in rows, response (dependent) and predictor (independent) variables in columns. For models with more than one independent (predictor) variable, the 2-D data table will encode the variables as multiple columns. This means that for modeling the data, each time an event is observed (one row), it must be assigned to the (many) correct values of the predictor variable columns. Since the event code is a single label, i.e., a 1 or “beep” or 11, dropping it into the variable columns does not correctly specify the model. Using logged event codes directly as predictor variables can only work in simple designs with one factor. It does not generalize to multiple predictor variables.
Furthermore, the relationship between logged integer event codes and predictor variables in models of real interest is rarely fully specified when the data are initially recorded. The sequence of events is not fixed when there is opportunity for human interaction as there is in any design which involves behavioral responses. For instance when the event code for a target stimulus is logged, there is no way to know whether it was detected (“hit”) or not (“miss) until a response is made later. This means the experimental variable “accuracy” cannot be read off the stimulus event code alone, it is contingent on a sequence of codes. Furthermore, the relevant factors and levels may not have been built into the integer codes in the first place. Perhaps your first hypothesis was a bust and you want to investigate other variables. Or Reviewer #2 has a pet theory and insists some variable be included. The integer event codes fossilized in the recording have to be recoded after the fact for the variables you or Reviewer 2 now want to model.
So besides the fact that event codes are simple labels not quantities, a further problem is that the time-stamped integer event codes are fixed once and for all when the data are first recorded but the variables associated with the event for modeling often are not. The obvious solution is a look-up table that maps the logged event codes to the appropriate values of the predictor variables. Though conceptually straightforward, there are a few complications in practice.
The variables associated with a specific event are often contingent on whether other events occur and if so when. The variables always depend on the model and there are always alternative models. The events may be initially coded for event type, e.g., codes 1 and 2 for congruous and incongruous words and the codes may not differentiate other relevant attributes of the events, e.g., differences in length, familiarity, age of acquisition of the individual words of the two types. This means that in practice, the form of the relationship (mapping) between individual fossilized event codes and the (levels of) categorical and (values of) continuous predictor variables included in a given model might be anything: one-to-one, one-to-many, many-to-one, or many-to_many.
code tagging¶
The code tagging approach automates arbitrary mappings from integer event code (sequences) to categorical and continous predictor variables for some common cases in designed experiments. The general framework is built around tidy tabular data structures which simplifies the implementation of custom re-coding for cases that are not handled by the automation.
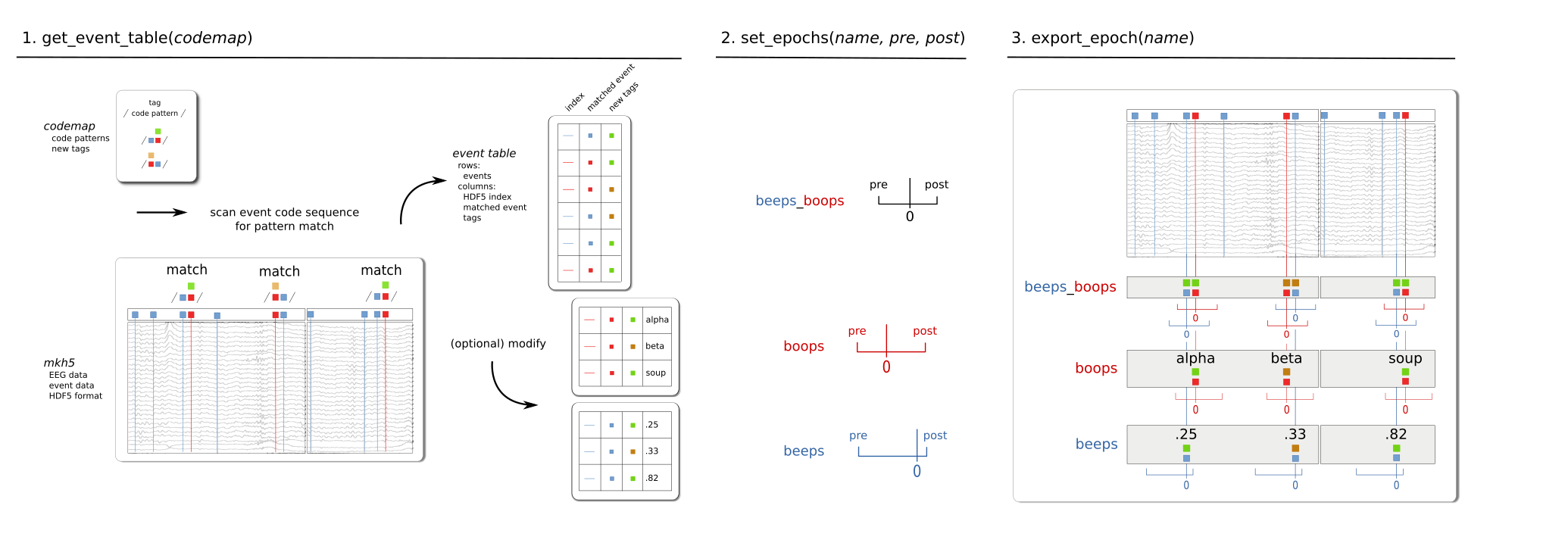
Three steps from EEG recordings to fixed-length EEG epochs tagged for modeling.¶
- codemap
The codemap is a recipe for re-coding (tagging) integer event codes with the additional categorical and continuous experimental variables (tags) as required for modeling real world experimental data in informative ways.
The codemap is a table (rows x columns) where each row is has the form, pattern tag, …, tag. This says, “any numerical event code (sequence) that matches this regular-expression-like pattern, gets these tags”.
The sample portion of a codemap shown next re-codes integer event 10 as a hit, miss, or one of two types of anticipation error of depending on what the response code is (1040, 1064) and whether it occurs after or before the code following the 10. It also tags 10 with .25, the numeric value of the continuous the continuous predictor variable “prob”, which gives the probability of the stimulus associated with code 10 in the stimlulus sequence. This is a stimulus attribute, i.e., not contingent on the response but potentially useful to encode for this and the rest of the stimuli if we are interested in the relationships between probability and accuracy and/or their impact on the EEG response variable we are modeling.
regexp
stim
accuracy
prob
(#10) \d+ 1040
beep
hit
.25
(#10) \d+ 1064
beep
miss
.25
(#10) 1040 \d+
beep
a_err1
.25
(#10) 1064 \d+
beep
a_err2
.25
The mapping between the patterns and the tags in a codemap is restricted only in that each pattern gets the same number of tags. The code patterns can be mapped to tags one-one, one-many, many-one, many-many as needed.
Depending on the specifics of the code pattern and the data, each code pattern may match the event data any number of times. A cooperative and attentive participant may not make any anticipation errors, others may.
Critically, the codemap is also an event filter. In the real experimental world, the event log is typicaly a record of many kinds of events. Some subsets of events are relevant for some analyses of the brain activity, other subsets for other analyses, and some are critically important for the experiment, but not directly relevant to the time course of brain activity, e.g., logging information about the sequence of trials and states of the stimulus delivery and data recording apparatus. A codemap dives into the overpopulated event stream and returns with a whitelist of all and only those events relevant for a particular analysis. Since analyses answer questions and, in general, there is more than one question that might be usefully asked of a given dataset, in general, there will be more than one codemap.
event table
Whereas the codemap is the recipe for linking events with experimental variables, the event table is the result of sweeping the code patterns across the recorded event data and collecting the matches in rows x columns. This creates a tidy, lightweight lookup table, where each row
shows exactly where to find the event in the digital data stream, i-th digital sample index.
the immutable event code as digitally recorded in the original data stream (plus other information relevant to for checking data integrity.)
and the highly and necessarily mutable categorical and continuous variables the analyst deem relevant for modeling brain activity in relation to the event which may be in many-many relations to the immutable event codes.
By design, the tabular rows=observations x columns=variables format of the event table is the same as the familiar observations x variables data format assumed by statistical modeling software like statsmodels and patsy in Python and lm and lme4 in R and can be broadcast from the event sample to other data samples, e.g., in a fixed length epoch. In the event table the digital event code is simply another tag for the event, one among many.
The rows and columns of the event table can and should be inspected to verify that codes were correctly matched and tagged.
Event tables may be further modified, e.g., by
pruning unnecessary rows (matched events) or columns (tags)
adding more tags (columns) from other data sources
adding more matched events (rows) by stacking or slicing the rows of other same-shape event tables from the same HDF5 file.
epochs
Epochs are fixed-length segments of the EEG and event recordings, time-stamped relative to a pattern-matched event code at time = 0 and tagged with the column variables from the event table. To get the epochs data for modeling, the information in the event table is stashed alongside the EEG recordings as an epochs table by providing a name and fixed-length epoch interval boundaries relative to the matched event at time 0 (pre, post). The EEG epochs can then be extracted from the HDF5 data file by name as a tidy table of indexed tagged tabular multi-channel time-series data. The epochs data can be read into the Python environment or exported as a separate file in various data interchange formats:
native HDF5 (h5)
pandas.DataFrame pytables HDF5Store (pdh5)
feather
For working in Python feather, h5, and pdh5 all work. The feather format is incrementally larger but much faster to read with pandas. For working in R feather is easiest, h5 also works. For working in MATLAB, h5 is easiest.
In the epochs data, the single trial EEG, the fossilized stimulus and response event codes, and the new stimulus and response variable tags all travel together. This makes it straightforward to verify event codes are tagged correctly and to synchronize single trial EEG and response measures and anlaysis.